| Journal of Clinical Medicine Research, ISSN 1918-3003 print, 1918-3011 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Clin Med Res and Elmer Press Inc |
| Journal website http://www.jocmr.org |
Short Communication
Volume 11, Number 6, June 2019, pages 458-463
Phenotyping to Facilitate Accrual for a Cardiovascular Intervention
Kavishwar B. Wagholikara, b, e, Christina M. Fischerc, Alyssa P. Goodsond, Christopher D. Herrickd, Taylor E. Macleanc, Katelyn V. Smithc, Liliana Ferac, Thomas A. Gazianoc, Jacqueline R. Dunningc, Joshua Bosque-Hamiltonc, Lina Mattac, Eloy Toscanod, Brent Richterd, Layne Ainsworthd, Michael F. Oatesd, Samuel Aronsond, Calum A. MacRaea, c, Benjamin M. Sciricaa, c, Akshay S. Desaia, c, Shawn N. Murphya, b
aHarvard Medical School, Boston, MA, USA
bMassachusetts General Hospital, Boston, MA, USA
cBrigham and Women’s Hospital, Boston, MA, USA
dPartners Healthcare, Boston, MA, USA
eCorresponding Author: Kavishwar B. Wagholikar, Harvard Medical School, Boston, MA, USA
Manuscript submitted April 2, 2019, accepted April 30, 2019
Short title: Phenotyping to Facilitate Accrual
doi: https://doi.org/10.14740/jocmr3830
| Abstract | ▴Top |
Background: The conventional approach for clinical studies is to identify a cohort of potentially eligible patients and then screen for enrollment. In an effort to reduce the cost and manual effort involved in the screening process, several studies have leveraged electronic health records (EHR) to refine cohorts to better match the eligibility criteria, which is referred to as phenotyping. We extend this approach to dynamically identify a cohort by repeating phenotyping in alternation with manual screening.
Methods: Our approach consists of multiple screen cycles. At the start of each cycle, the phenotyping algorithm is used to identify eligible patients from the EHR, creating an ordered list such that patients that are most likely eligible are listed first. This list is then manually screened, and the results are analyzed to improve the phenotyping for the next cycle. We describe the preliminary results and challenges in the implementation of this approach for an intervention study on heart failure.
Results: A total of 1,022 patients were screened, with 223 (23%) of patients being found eligible for enrollment into the intervention study. The iterative approach improved the phenotyping in each screening cycle. Without an iterative approach, the positive screening rate (PSR) was expected to dip below the 20% measured in the first cycle; however, the cyclical approach increased the PSR to 23%.
Conclusions: Our study demonstrates that dynamic phenotyping can facilitate recruitment for prospective clinical study. Future directions include improved informatics infrastructure and governance policies to enable real-time updates to research repositories, tooling for EHR annotation, and methodologies to reduce human annotation.
Keywords: Electronic health records; Intervention; Phenotyping; Cohort identification
| Introduction | ▴Top |
The adoption of electronic health records (EHRs) in the USA has generated large datasets that enable observational research. An essential step in utilizing these datasets for research is to identify patients with certain characteristics that match a set of eligibility criteria, a process known as phenotyping or patient cohort identification. Phenotyping requires significant manual effort to create labeled/validation datasets, and a population database is required to execute the algorithm [1].
Phenotyping has been reported to be useful for retrospective studies [2-4]; however, it has been relatively under-utilized for prospective studies. Although there has been considerable thought on phenotyping strategies for pragmatic clinical trials (PCTs) [5-7], much of the phenotyping for clinical studies has focused on developing disease registries or disease cohorts that serve as an initial focused pool for studies related to a particular disease [1, 8, 9].
Disease registries often cannot be readily used for prospective studies, as each study has specific eligibility criteria, and additional algorithms are thus necessary to implement the eligibility criteria to refine the disease registry, which costs significant effort. Due to the high cost and effort involved, phenotyping has largely been used for pre-screening before starting recruitment, and no previous reports of actively phenotyping for intra-study recruitment have been published to date. To address this gap, we describe an approach for phenotyping in tandem with manual screening, referred to as dynamic phenotyping (DP). We implement this approach to facilitate recruitment for a prospective study on heart failure.
The study team had utilized a phenotyping algorithm to identify an initial cohort of heart failure patients prior to commencing recruitment. However, as the study progressed, the study team recognized the need to refine the disease-cohort to match the eligibility criteria, which evolved into an approach of dynamically performing phenotyping in tandem with manual screening. We report on the challenges encountered when using this approach, and we test the hypothesis that DP can significantly improve the screening rate.
| Materials and Methods | ▴Top |
This study was carried out at Partners Healthcare, Boston and was approved by the Institutional Review Board. For the case study, we performed phenotyping using data for 193,808 patients with a coded diagnosis of heart failure. This study was conducted in compliance with the ethical standards of the responsible institution on human subjects as well as with the Helsinki Declaration.
DP
Our approach consists of multiple screen cycles. At the start of each cycle, a phenotyping algorithm is used to identify eligible patients from the EHR, creating an ordered list such in which those patients most likely to be eligible are listed first. This list is manually screened, and the results are analyzed to improve phenotyping for the next cycle. We applied this approach to EHR data at Brigham and Women’s Hospital (BWH) to accrue heart failure patients with reduced ejection fraction (HFrEF) for an intervention study, as described below in the methods section.
Heart failure study
Although national guidelines exist for the pharmacological therapy of HFrEF, many eligible patients do not receive optimal guideline-directed medical therapy. To address this gap, we created a program to remotely optimize HFrEF patient care using supervised non-licensed navigators. DP was used to identify patients eligible for the study. Treating providers were approached for consent to adjust medical therapy in accordance with ACC/AHA guidelines [10, 11]. Once consent was obtained, patients were contacted by phone by a navigator who completes a medication reconciliation and provides education over the telephone [28-32].
We obtained data for 193,808 patients with a coded diagnosis of heart failure from the institutional research patient data repository [12-15]. The eligibility criteria for the study included adult patients that had a diagnosis of HFrEF, and whose cardiac care was managed primarily by a cardiologist at BWH. Patients were excluded for the following reasons: end-stage renal disease, active chemotherapy, low life expectancy, risk of frailty, transplant, intravenous inotrope use, prior need for mechanical ventricular support, unstable disease, and/or a history of medication non-adherence.
The primary challenge was to identify eligible patients from the pool of patients that had a coded diagnosis of heart failure. As the distributions for exclusion criteria were expected to be small, we focused on implementing the inclusion criteria through in-silico phenotyping.
We utilized a variety of approaches to develop algorithms for extracting variables (Table 1), including rules, regular expressions for text processing and logistic regression [16]. In addition, we measured the performance of phenotyping in each cycle by measuring the positive screen rate (PSR) on the annotations resulting from manual screening in the next cycle.
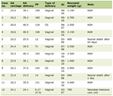 Click to view | Table 1. List of Algorithms for Pre-Screening |
| Results | ▴Top |
We applied the DP approach to facilitate recruitment for a real-world study on in HFrEF. Table 2 presents the results of the first four cycles, while Figure 1 summarizes the results for each cycle.
 Click to view | Table 2. PSR in Each Screening Cycle |
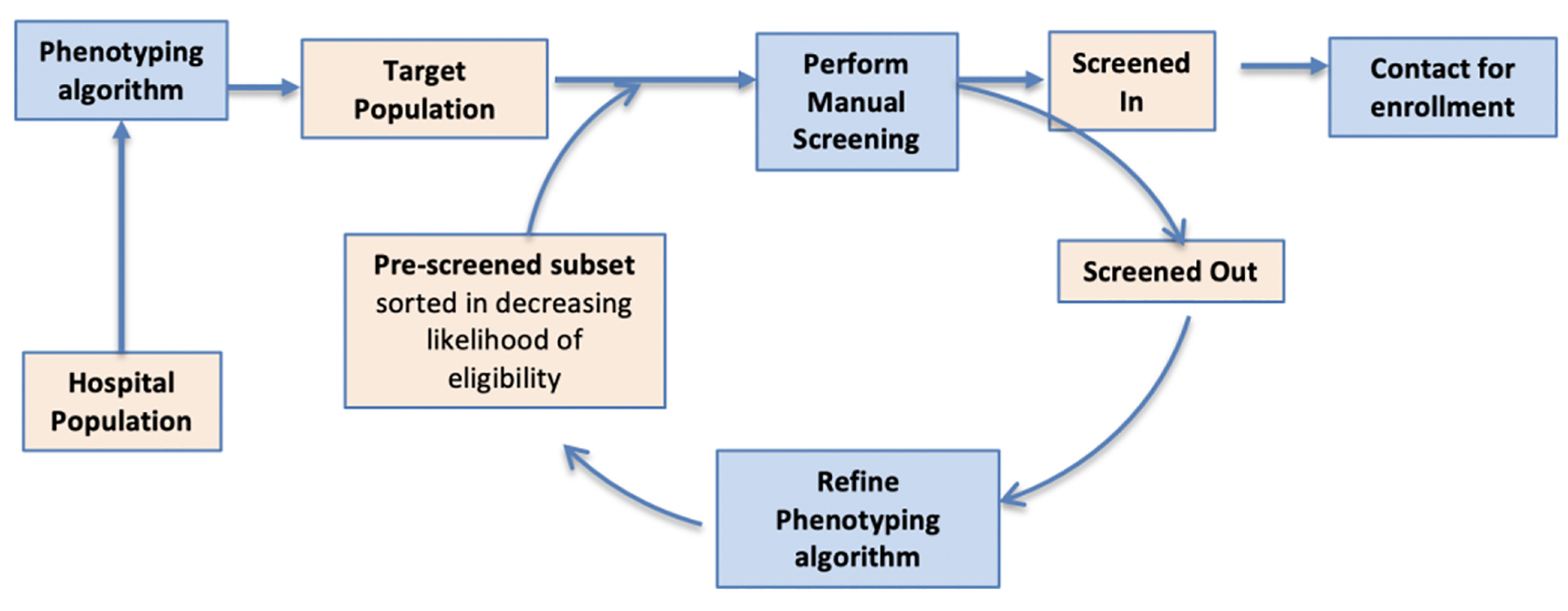 Click for large image | Figure 1. DP consists of multiple screen cycles. At the start of each cycle, phenotyping is used to identify eligible patients from the EHR, creating an ordered list in which the most eligible patients are listed first. This list is manually screened, and the results are analyzed to improve phenotyping for the next cycle. DP: dynamic phenotyping; EHR: electronic health record. |
Cycle I
The initial cohort was developed using: 1) a logistic regression model trained on a dataset of 250 patient records that were manually annotated for heart failure, 2) a regular expression for extracting ejection fraction (EF), and 3) a rule that the care provider with the highest count of cardiology notes was identified as the primary cardiologist.
Major reasons for ineligibility included high EF, incorrect identification of primary cardiologist and no heart failure. Half of the false positives were due to a combination of incorrect extraction of EF, heart failure and incorrect identification of primary cardiologist, which motivated the study team to refine the phenotyping algorithm.
Cycle II
We improved the algorithm extraction of EF by developing patterns to identify echocardiograms and by using regular expressions to extract numbers, ranges, and prose mentions of EF. This approach was found to eliminate false positives due to incorrect EF extraction. Moreover, accurate EF extraction obviated the need for the heart failure algorithm, since, by definition, EF ≤ 40 is logically sufficient to enter the intervention program.
To improve identification of the primary cardiologist, we added the restriction of having two visits to the institutional cardiologist within a 2-year period, a finding that was suggested by the navigation team. However, as observed in Figure 2, this restriction did not decrease the number of false positives.
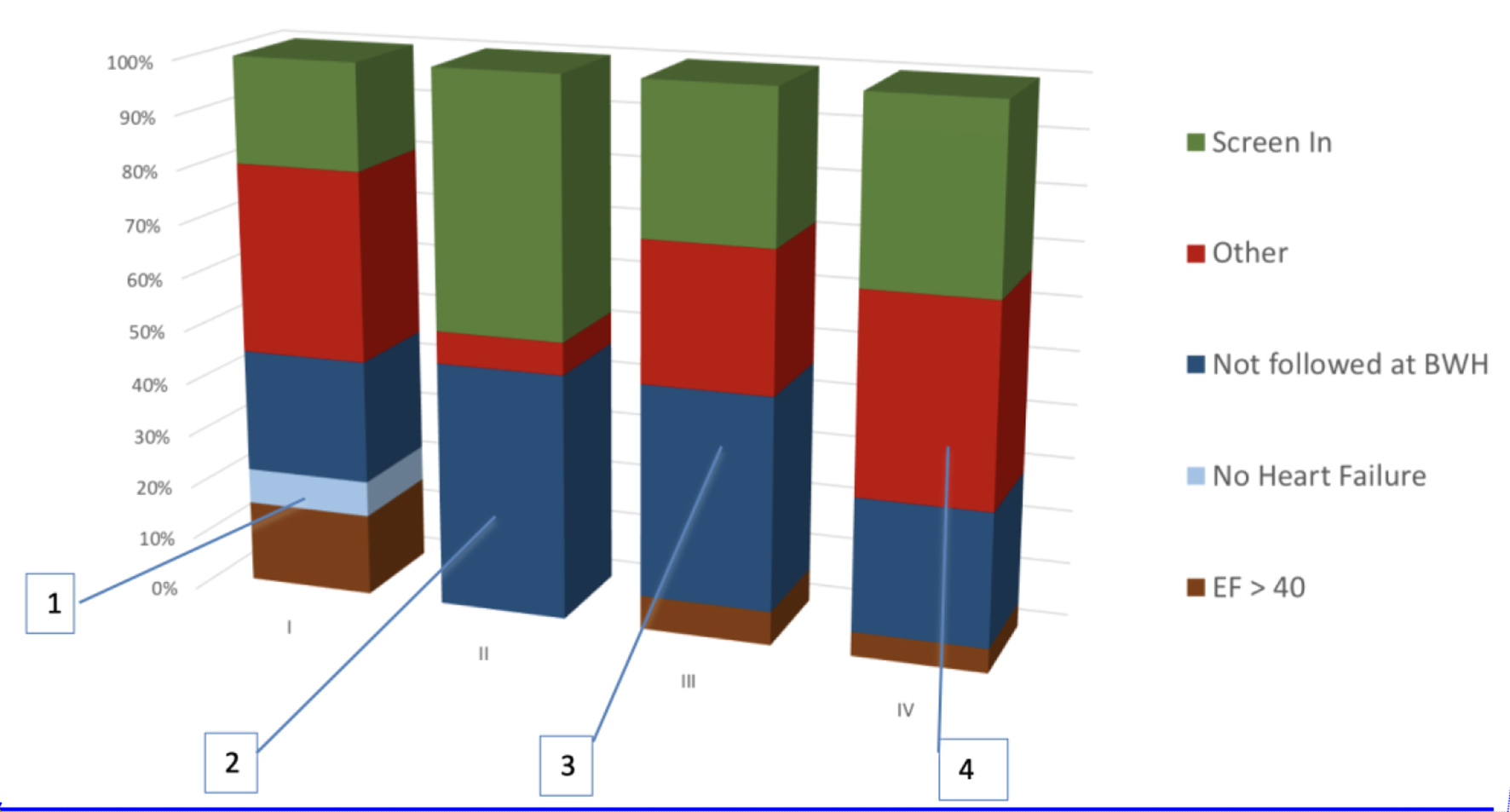 Click for large image | Figure 2. Composition of manual screening performed in each cycle. 1) No HF and EF accounted for 25% of false positives. HF was detected by machine learning and EF was extracted using a simple regular expression from clinical notes. 2) Optimization of EF parser and inferring HF from low EF eliminated many false positives. But failure to exclude patients not primarily managed by the outpatient cardiology clinic at BWH emerged as a challenge. This was because the cardiologist was inferred using total number of EHRs entries authored for the patient. 3) Cardiologist was inferred from number of EHR entries limited to the outpatient setting. But this did not significantly reduce false positives. 4) Use of machine learning to infer the primary cardiologist significantly excluded patients that are not managed at BWH. HF: heart failure; EF: ejection fraction; BWH: Brigham and Women’s Hospital; EHR: electronic health record. |
Cycle III
We further attempted to improve detection of the cardiologist by using the total number of EHRs authored by the cardiologists. False positives due to EF extraction errors reappeared in this cycle due to a lag in updates to the research repository; the patients had undergone echocardiography after the repository update.
Cycle IV
The limited performance of rule-based approaches for identifying the primary cardiologist led the study team to develop a logistic regression-based model, which was found to reduce false positives (Fig. 2).
PSR
The PSR improved in cycles II-IV over the first cycle, and the iterative approach resulted in an improvement in PSR; the P-value of the Chi-square statistic upon comparing average PSR for cycles II-IV (36%) and to PSR for the first cycle (20%) is < 0.01.
| Discussion | ▴Top |
A total of 1,022 patients were screened, with 223 (23%) of patients being found eligible for enrollment into the intervention program. When a single run of phenotyping is used, the pool of eligible patients is expected to decline as manual screen progresses, resulting in more false positives and a subsequently downward trend in the PSR. We used iteratively improved phenotyping in each screening cycle. This process offset the dip in the PSR by providing an enriched cohort in every cycle. Without an iterative approach, the PSR was expected to dip below the 20% measured in the first cycle; however, the cyclical approach increased the PSR to 23%.
Recruitment for prospective clinical studies has been reported to be resource intensive requiring a large number of patients to be screened in order to achieve enrollment goals [1, 17-27]. Hence, DP may be useful to improve the screening rate for clinical studies.
One advantage of the dynamic approach is that manual screening performed by the navigators in each cycle provides a better perspective on the distribution of false positives. The latter helps direct the focus of the phenotyping team for refining the algorithms for the next cycle. However, one caveat is the additional resources required for phenotyping to keep up with the rapid operational pace of the clinical study.
A major challenge for DP is the lack of research informatics infrastructure necessary to maintain the performance and real-time data needs for such analyses. Specifically, the gaps are the latency in updates to research repositories from the EHR and lack of tooling for navigators to record annotations while performing manual screening. The latency in updates to research repositories lags behind EHR updates by days to weeks. Hence, patients selected by phenotyping often have events and visits during the lag period, which leads to false positives. For instance, the false positives in cycles III and IV due to incorrect EF were a result of patients having new echocardiograms after an update to the research repository. While it is true that most research or care innovation studies do not require the most recent patient data, DP for prospective studies presents a new use-case that requires a near real-time repository [28].
The second infrastructure barrier was a lack of tooling that allows navigators to store their annotations for chart reviews. Instead, navigators used spreadsheet software to record their annotations, which required substantial curation for re-integration into the subsequent DP cycle.
Data governance also poses a challenge for DP, as researchers require the same data fields to develop algorithms that the navigators have access to in the EHR system. For instance, our research repository lacked the metadata of “clinical service type” for notes, and there was no encounter metadata for notes, both of which were available for navigators that helped researchers identify the primary cardiologist for the patient. The absence of these fields in our research repository resulted in additional effort being necessary for the development of the phenotyping algorithm.
Furthermore, we terminated the manual screening phase (in the DP cycles) when the PSR was below a subjective threshold. The manual screening was often prolonged due to logistical reasons such as delays in the updating of the research repository for executing the phenotyping. In an ideal scenario, DP cycles are executed in real-time and the phenotyping is executed after each manual screening/annotation.
Limitations
One limitation in our study was that we did not extensively investigate optimization of the phenotyping algorithms. Secondly, we did not implement a model for exclusion criteria, which forms a significant proportion of false positives (Fig. 2). Both of these limitations were due to the required fast pace of the study; in retrospect, it would have further improved the PSR. Also, the phenotyping algorithms may be converged more rapidly by utilizing active learning and semi-supervised approaches [29-31].
Conclusions
Our study demonstrates that DP can facilitate recruitment for prospective clinical study. Future directions include improved informatics infrastructure and governance policies to enable real-time updates to research repositories, tooling for EHR annotation and methodologies to reduce human annotation.
Acknowledgments
None.
Financial Disclosure
This study was supported by NIH grants R01-HG009174 and R00-LM011575, and funding from Partner Healthcare.
Conflict of Interest
The authors have no conflict of interest to disclose.
Informed Consent
The institutional review board approved a waiver of informed consent.
Author Contributions
The phenotyping algorithms were developed by KBW, CMF, AG, CH, LA and MO, and received assistance from MR, ET and BR. CMF led the navigation team for manual screening, which included TEM, KVS, LES, TAG, JRD, JBH and LM. The case study was envisioned by CAM, BMS, ASD and SNM. All authors contributed to the manuscript and approved the final version.
| References | ▴Top |
- Geraci J, Wilansky P, de Luca V, Roy A, Kennedy JL, Strauss J. Applying deep neural networks to unstructured text notes in electronic medical records for phenotyping youth depression. Evid Based Ment Health. 2017;20(3):83-87.
doi pubmed - Warrer P, Hansen EH, Juhl-Jensen L, Aagaard L. Using text-mining techniques in electronic patient records to identify ADRs from medicine use. Br J Clin Pharmacol. 2012;73(5):674-684.
doi pubmed - Wagholikar KB, MacLaughlin KL, Chute CG, Greenes RA, Liu H, Chaudhry R. Granular quality reporting for cervical cytology testing. AMIA Jt Summits Transl Sci Proc. 2015;2015:178-182.
- Kaelber DC, Foster W, Gilder J, Love TE, Jain AK. Patient characteristics associated with venous thromboembolic events: a cohort study using pooled electronic health record data. J Am Med Inform Assoc. 2012;19(6):965-972.
doi pubmed - Richesson RL, Green BB, Laws R, Puro J, Kahn MG, Bauck A, Smerek M, et al. Pragmatic (trial) informatics: a perspective from the NIH Health Care Systems Research Collaboratory. J Am Med Inform Assoc. 2017;24(5):996-1001.
doi pubmed - Richesson RL, Hammond WE, Nahm M, Wixted D, Simon GE, Robinson JG, Bauck AE, et al. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory. J Am Med Inform Assoc. 2013;20(e2):e226-231.
doi pubmed - Weinfurt KP, Hernandez AF, Coronado GD, DeBar LL, Dember LM, Green BB, Heagerty PJ, et al. Pragmatic clinical trials embedded in healthcare systems: generalizable lessons from the NIH Collaboratory. BMC Med Res Methodol. 2017;17(1):144.
doi pubmed - Kagawa R, Kawazoe Y, Ida Y, Shinohara E, Tanaka K, Imai T, Ohe K. Development of Type 2 Diabetes Mellitus Phenotyping Framework Using Expert Knowledge and Machine Learning Approach. J Diabetes Sci Technol. 2017;11(4):791-799.
doi pubmed - Wood WA, Bennett AV, Basch E. Emerging uses of patient generated health data in clinical research. Mol Oncol. 2015;9(5):1018-1024.
doi pubmed - Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Jr., Colvin MM, Drazner MH, et al. 2017 ACC/AHA/HFSA Focused Update of the 2013 ACCF/AHA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Failure Society of America. J Card Fail. 2017;23(8):628-651.
doi pubmed - Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Jr., Drazner MH, Fonarow GC, et al. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2013;62(16):e147-239.
- Murphy SN, Avillach P, Bellazzi R, Phillips L, Gabetta M, Eran A, McDuffie MT, et al. Combining clinical and genomics queries using i2b2 - Three methods. PLoS One. 2017;12(4):e0172187.
doi pubmed - Thiemann VS, Xu T, Rohrig R, Majeed RW. Automated report generation for research data repositories: from i2b2 to PDF. Stud Health Technol Inform. 2017;245:1289.
- Wagholikar KB, Mandel JC, Klann JG, Wattanasin N, Mendis M, Chute CG, Mandl KD, et al. SMART-on-FHIR implemented over i2b2. J Am Med Inform Assoc. 2017;24(2):398-402.
doi pubmed - Wagholikar KB, Mendis M, Dessai P, Sanz J, Law S, Gilson M, Sanders S, et al. Automating installation of the Integrating Biology and the Bedside (i2b2) platform. Biomed Inform Insights. 2018;10:1178222618777749.
doi pubmed - Wagholikar KB, Fischer CM, Goodson A, Herrick CD, Rees M, Toscano E, MacRae CA, et al. Extraction of Ejection Fraction from Echocardiography Notes for Constructing a Cohort of Patients having Heart Failure with reduced Ejection Fraction (HFrEF). J Med Syst. 2018;42(11):209.
doi pubmed - Cheng SK, Dietrich MS, Dilts DM. A sense of urgency: Evaluating the link between clinical trial development time and the accrual performance of cancer therapy evaluation program (NCI-CTEP) sponsored studies. Clin Cancer Res. 2010;16(22):5557-5563.
doi pubmed - Lara PN, Jr., Higdon R, Lim N, Kwan K, Tanaka M, Lau DH, Wun T, et al. Prospective evaluation of cancer clinical trial accrual patterns: identifying potential barriers to enrollment. J Clin Oncol. 2001;19(6):1728-1733.
doi pubmed - Mannel RS, Walker JL, Gould N, Scribner DR, Jr., Kamelle S, Tillmanns T, McMeekin DS, et al. Impact of individual physicians on enrollment of patients into clinical trials. Am J Clin Oncol. 2003;26(2):171-173.
doi pubmed - Rahman M, Morita S, Fukui T, Sakamoto J. Physicians' reasons for not entering their patients in a randomized controlled trial in Japan. Tohoku J Exp Med. 2004;203(2):105-109.
doi pubmed - Siminoff LA, Zhang A, Colabianchi N, Sturm CM, Shen Q. Factors that predict the referral of breast cancer patients onto clinical trials by their surgeons and medical oncologists. J Clin Oncol. 2000;18(6):1203-1211.
doi pubmed - Welch BM, Marshall E, Qanungo S, Aziz A, Laken M, Lenert L, Obeid J. Teleconsent: A Novel Approach to Obtain Informed Consent for Research. Contemp Clin Trials Commun. 2016;3:74-79.
doi pubmed - Ghebre RG, Jones LA, Wenzel JA, Martin MY, Durant RW, Ford JG. State-of-the-science of patient navigation as a strategy for enhancing minority clinical trial accrual. Cancer. 2014;120(Suppl 7):1122-1130.
doi pubmed - Renfro LA, Sargent DJ. Statistical controversies in clinical research: basket trials, umbrella trials, and other master protocols: a review and examples. Ann Oncol. 2017;28(1):34-43.
doi pubmed - Afrin LB, Oates JC, Kamen DL. Improving clinical trial accrual by streamlining the referral process. Int J Med Inform. 2015;84(1):15-23.
doi pubmed - Embi PJ, Jain A, Clark J, Harris CM. Development of an electronic health record-based Clinical Trial Alert system to enhance recruitment at the point of care. AMIA Annu Symp Proc. 2005;2005:231-235.
pubmed - Carlson RW, Tu SW, Lane NM, Lai TL, Kemper CA, Musen MA, Shortliffe EH. Computer-based screening of patients with HIV/AIDS for clinical-trial eligibility. Online J Curr Clin Trials. 1995;Doc No 179[3347 words; 3332 paragraphs].
- Wagholikar KB, Jain R, Oliveira E, Mandel J, Klann J, Colas R, Patil P, et al. Evolving research data sharing networks to clinical App sharing networks. AMIA Jt Summits Transl Sci Proc. 2017;2017:302-307.
- Yu S, Chakrabortty A, Liao KP, Cai T, Ananthakrishnan AN, Gainer VS, Churchill SE, et al. Surrogate-assisted feature extraction for high-throughput phenotyping. J Am Med Inform Assoc. 2017;24(e1):e143-e149.
- Yu S, Liao KP, Shaw SY, Gainer VS, Churchill SE, Szolovits P, Murphy SN, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc. 2015;22(5):993-1000.
doi pubmed - Yu S, Ma Y, Gronsbell J, Cai T, Ananthakrishnan AN, Gainer VS, Churchill SE, et al. Enabling phenotypic big data with PheNorm. J Am Med Inform Assoc. 2018;25(1):54-60.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Clinical Medicine Research is published by Elmer Press Inc.


